Note
Go to the end to download the full example code
Quick-Start: Running a simple EMD#
This getting started tutorial shows how to use EMD to analyse a synthetic signal.
Running an EMD and frequency transform#
First of all, we import both the numpy and EMD modules:
# sphinx_gallery_thumbnail_number = 2
import matplotlib.pyplot as plt
import numpy as np
import emd
We then define a simulated waveform containing a non-linear wave at 5Hz and a sinusoid at 1Hz:
sample_rate = 1000
seconds = 10
num_samples = sample_rate*seconds
time_vect = np.linspace(0, seconds, num_samples)
freq = 5
# Change extent of deformation from sinusoidal shape [-1 to 1]
nonlinearity_deg = 0.25
# Change left-right skew of deformation [-pi to pi]
nonlinearity_phi = -np.pi/4
# Compute the signal
# Create a non-linear oscillation
x = emd.simulate.abreu2010(freq, nonlinearity_deg, nonlinearity_phi, sample_rate, seconds)
x += np.cos(2 * np.pi * 1 * time_vect) # Add a simple 1Hz sinusoid
x -= np.sin(2 * np.pi * 2.2e-1 * time_vect) # Add part of a very slow cycle as a trend
# Visualise the time-series for analysis
plt.figure(figsize=(12, 4))
plt.plot(x)
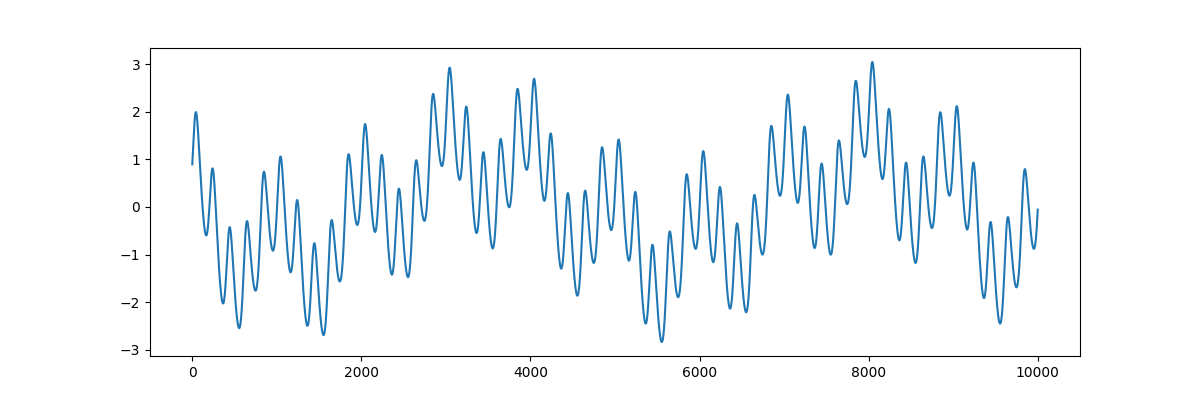
[<matplotlib.lines.Line2D object at 0x7f0ec7ba3d90>]
Try changing the values of nonlinearity_deg and nonlinearity_phi to
create different non-sinusoidal waveform shapes.
Next, we can then estimate the IMFs for the signal:
imf = emd.sift.sift(x)
print(imf.shape)
(10000, 4)
and, from the IMFs, compute the instantaneous frequency, phase and amplitude using the Normalised Hilbert Transform Method:
IP, IF, IA = emd.spectra.frequency_transform(imf, sample_rate, 'hilbert')
From the instantaneous frequency and amplitude, we can compute the Hilbert-Huang spectrum:
# Define frequency range (low_freq, high_freq, nsteps, spacing)
freq_range = (0.1, 10, 80, 'log')
f, hht = emd.spectra.hilberthuang(IF, IA, freq_range, sum_time=False)
Visualising the results#
we can now plot some summary information, first the IMFs:
emd.plotting.plot_imfs(imf)
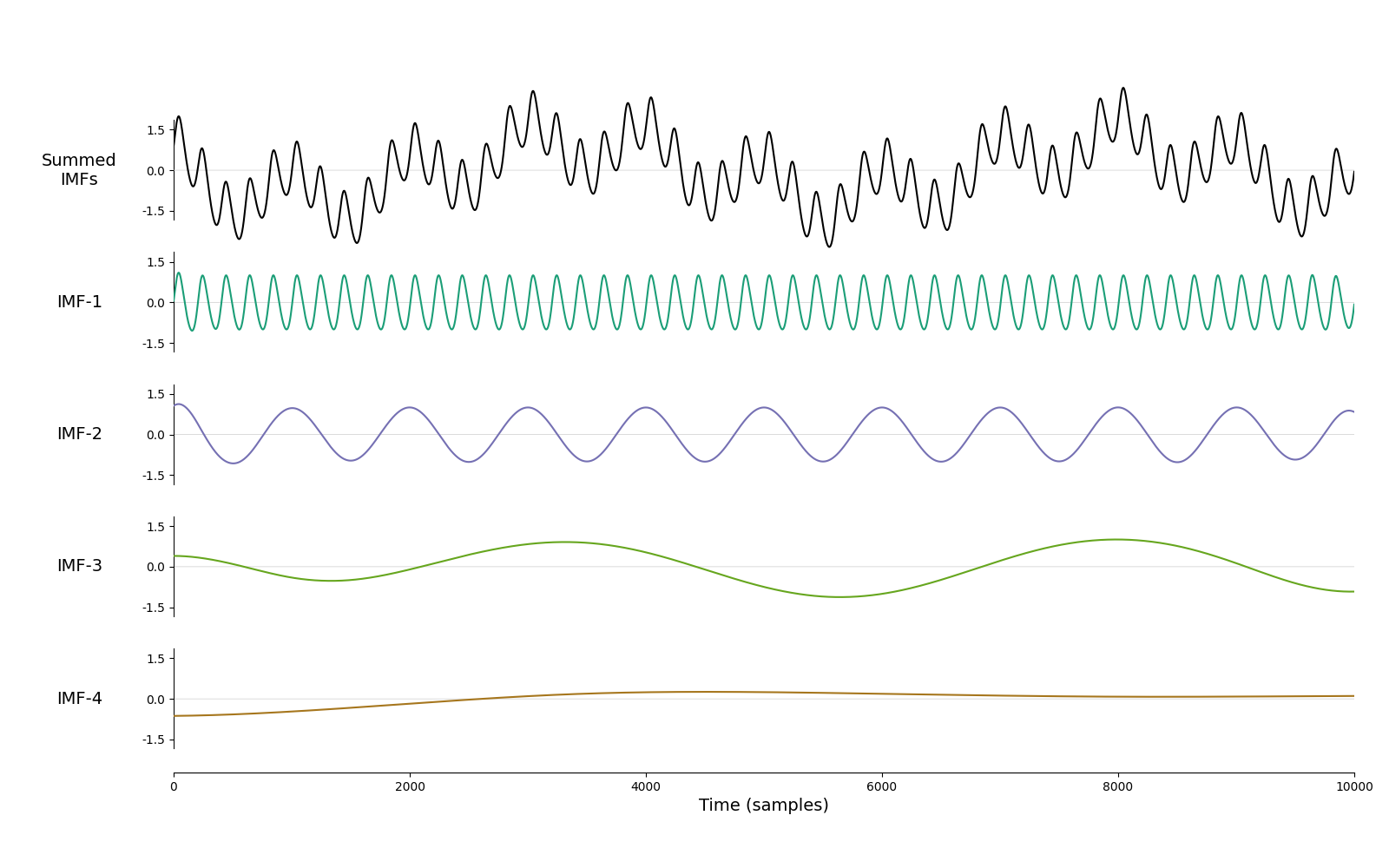
<Axes: xlabel='Time (samples)'>
and now the Hilbert-Huang transform of this decomposition
fig = plt.figure(figsize=(10, 6))
emd.plotting.plot_hilberthuang(hht, time_vect, f,
time_lims=(2, 4), freq_lims=(0.1, 15),
fig=fig, log_y=True)
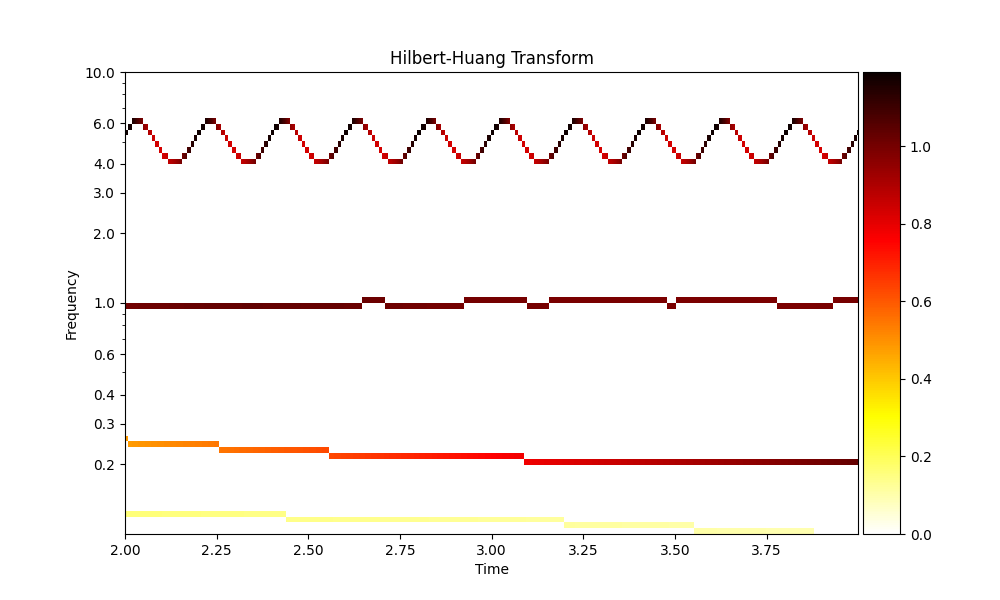
<Axes: title={'center': 'Hilbert-Huang Transform'}, xlabel='Time', ylabel='Frequency'>
Total running time of the script: (0 minutes 1.581 seconds)